Wallin, A.E., Salmi, A., Tuma, R. (2007). Step Length Measurement--Theory and Simulation for Tethered Bead Constant-Force Single Molecule Assay. Biophysical Journal, 93(3), 795-805. DOI: 10.1529/biophysj.106.097915
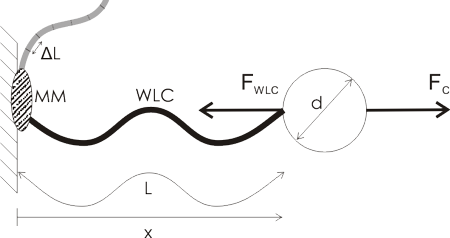
A lot of single molecule experiments are carried out in the configuration shown above. The idea is to mechanically probe a bio-molecule (such as DNA or RNA) or find out how an enzyme (such as RNAP) or a molecular motor (e.g. viral packaging motor) works. You either optically or magnetically trap a microsphere (the round thing with diameter d), which allows you to move it around, measure its position very accurately, and measure the forces acting on the sphere. The sphere is a big thing, usually 500 to 1000 nm in diameter, so it's easily visible in the microscope (the molecules themselves are not visible unless you label them with fluorescent tags).
Then you ask a biochemist in your lab to bind some useful stuff to the sphere. I've illustrated a hypothetical experiment where a molecular motor (MM, grey blob) is bound to the sample chamber wall, and the bead is tethered to the motor via a Worm-Like Chain (WLC, thick wavy line, a common model for polymers such as DNA/RNA).
The problem is that these things are tiny! People want to measure single steps for molecular motors, which can be as short as 0.3 nm. At the same time biological things also live at close to room temperature, so both the molecules and the microsphere are affected by random collisions from the solvent molecules.
Now the question is how small steps or changes in the WLC length are really measurable? It's a bit like going on a walk with your dog: imagine a very flexible string between the dog and yourself. Now if you pull only weakly on the string you can't expect to feel very much of the individual steps the dog takes, you just follow along smoothly while keeping the force approximately constant. If you really want to feel the individual steps you would use a stiffer string and pull much harder on it(but the dog would not like that...). Previous work such as that by Gittes and Schmidt and Moffitt et al. pretty much agrees that the Signal-to-Noise-Ratio (SNR) should look something like this:
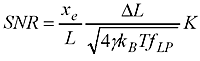
where you've used a force to stretch out the WLC (of length L) to an extension xe, the steps you are trying to detect are of size DeltaL, and the stiffness of the WLC is K. In the square root we have gamma (Stokes drag coefficient on the bead), Boltzmann's constant kb, temperature T, and f_lp the measurement bandwidth.
What do we need to do to get a high SNR (and thus measure short steps accurately) ? Use a large force (that increases xe and K), use small beads (that decreases gamma), cool down the lab (decrease T, but not so much so that your biology dies!), and measure slow moving stuff (low f_lp). But this formula is not so practical to use. You need to work out the extension xe yourself and also estimate the stiffness K somehow. I couldn't find anyone who had addressed how this works for common tethers such as DNA and RNA.
My (small) contribution to these thoughts is mainly working out how the SNR really depends on the properties of the WLC and the pulling force. I did a bit of simple theoretical analysis and showed with a Monte-Carlo simulation that the theoretical prediction is about right. This results in SNR 'maps' in parameter space that hopefully can guide experiments. For example if you suspect that your enzyme has a certain step length and you know your tether properties you can use the map or an approximate formula to work out the force you need to have a chance of measuring individual steps.
A recent review by Moffitt et al. from the Bustamante lab makes some very similar points.
http://dx.doi.org/10.1146/annurev.biochem.77.043007.090225
in particular their Figure 2b is similar to the ones I made, indicating in a (step rate, step length) diagram what kind of experiments are likely to detect single steps and what experiments will likely not detect steps.
The paper is now freely available at PubMed:
http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1913155
cited by this paper
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2777445/?report=abstract