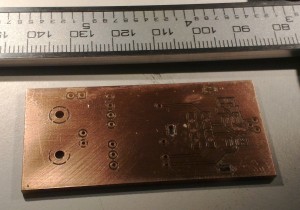
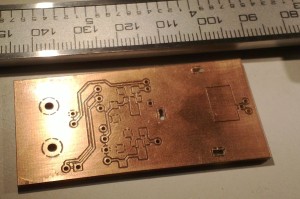
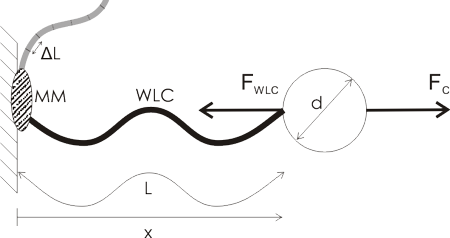
I'm making photodiode (transmipedance) amplifiers, and here is the first PCB being milled today. In the foreground a test-run where the cutter-height was too low resulting in too thin or vanishing PCB-traces. Note how the PCB material is not held in place along the Z-axis at all. The PCB-blank is just located in X/Y on the table using two locating pins/holes. In the Z-direction the idea is that the pneumatic cylinder pushes the lower flange of the spindle into contact with the PCB-material, and the exact cutter-height is adjusted relative to this flange only.
The toolchain is (old!) commercial software: PADS PowerLogic for schematic design, PADS PowerPCB for PCB-design, CircuitCam for converting the gerbers to HPGL, which BoardMaster uses to drive the mill (over RS232).
For general purpose 3D CAD at work we have Vertex (a Finnish Inventor/SolidWorks clone) and I used it to draw a model of the amplifier:
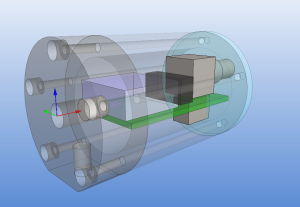
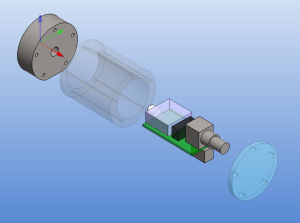
The size of the PCB and enclosure is mostly limited by how much of the powersupply one wants on-board, and how big connectors one wants to use. I'm using a standard BNC connector (SMA would have been smaller). The board is powered by a +9...18VDC supply which is DC2DC converted into +/-12 V and then regulated to +/- 5 V for the op-amp circuit. The box at the front is an RF shield for the amplifier itself. Light enters through an 8 mm hole in the face-plate and hits a TO-18 mounted photodiode. More on the circuit later.
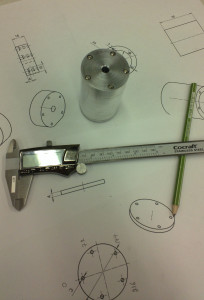
The enclosure is 48 mm in diameter with a 16 mm thick face-plate, a 4 mm thick back-plate, and the body (55 mm length) bored out to an inner-diameter of 34 mm. The body should fit a 25x54mm PCB. The end-plates are attached to the body with five M3 screws on a 40 mm diameter bolt-circle. There is an M6 thread on the bottom of the face-plate, for attaching the amplifier to an optical-table or other instrumentation. I made two of these from 50 mm aluminium round-bar on a manual lathe and mill (using a rotary table for the holes/threads).
Note: for manual machining five evenly spaced holes the angle-sequence is: 0 - 72 - 144 - 216 - 288 - 0.
I'm thinking about polishing these a bit and then anodizing them. But for RF-shielding the contact-surfaces of all three parts would then have to be sanded/milled-down after andoizing. to ensure good electrical conductivity between the parts.